Plasmids diversity and carbapenem-resistant Enterobacteriaceae outbreaks in a Swiss hospital
Antibiotics resistance vectors
We investigated the spread of carbapenem-resistant Enterobacteriaceae (CRE), a major public health concern, in a Swiss hospital. These bacteria, particularly Klebsiella pneumoniae and Serratia marcescens, pose a severe threat as they are resistant to last-resort antibiotics. Hospitals are hotspots for outbreaks due to environmental reservoirs like sink traps, which can harbor resistant bacteria and facilitate their transmission. The study underscores the importance of understanding the genetic mechanisms, particularly plasmids carrying resistance genes, to devise effective infection control strategies.

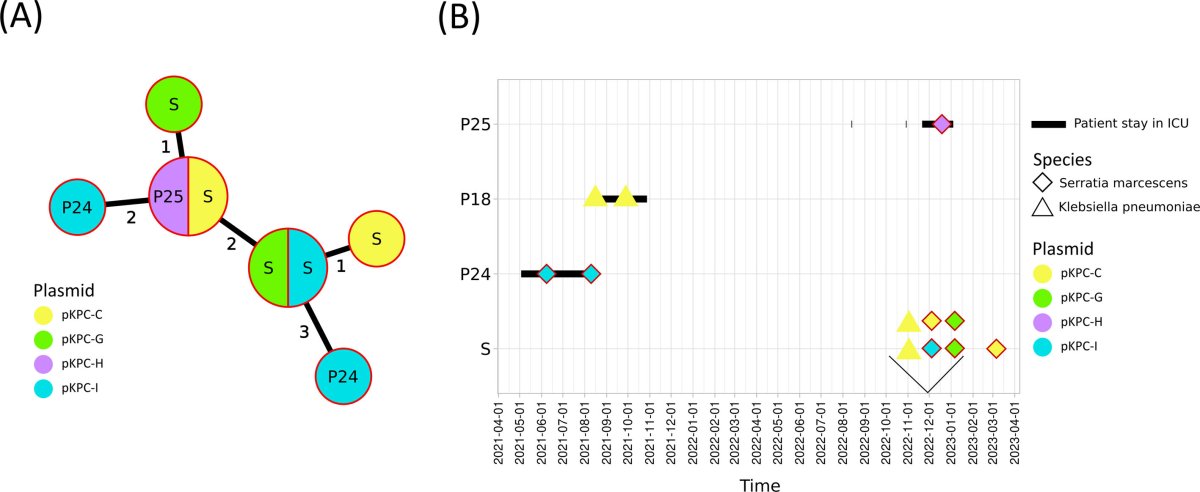
Over two years, we analyzed 57 isolates from patients and sink traps during two CRE outbreaks. Using advanced DNA sequencing techniques, they identified key resistance genes (blaKPC-2 and blaNDM-1) and characterized the plasmids carrying them. The study revealed a remarkable diversity of plasmids, showing that they spread not only within specific bacterial clones but also across different bacterial species. This indicates that sink traps serve as persistent reservoirs, facilitating plasmid exchange. Notably, plasmids were sometimes more varied than the bacteria themselves, suggesting that resistance spreads independently of bacterial clonal expansion.
These findings highlight the role of environmental reservoirs, like sink traps, in maintaining and spreading antibiotic resistance. This challenges traditional outbreak definitions that focus solely on bacterial clones. Effective infection control must consider both the bacterial strains and the plasmids driving resistance. The study advocates for integrated surveillance and comprehensive infection prevention measures to mitigate the spread of CRE in healthcare settings.
For more information, see the published paper.