Genetic markers for distinguishing individuals in mixed blood samples
Tackling the challenges of complex forensic samples
Forensic DNA analysis often encounters challenges in interpreting mixed samples where DNA from multiple contributors is present, especially when one donor's contribution is minuscule. This study tackles this problem by exploring and validating novel genetic markers called DIP-STRs (Deletion/Insertion Polymorphisms paired with Short Tandem Repeats). These markers excel in identifying minor DNA contributors in highly unbalanced mixtures, making them vital for forensic applications. By leveraging whole-genome sequencing data, this research contributes to advancing forensic genetics tools, enabling more precise and sensitive DNA profiling techniques.

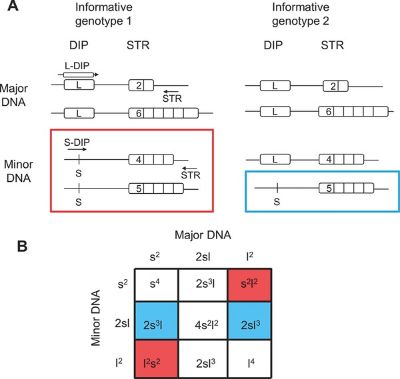
The study utilized data from over 76,000 human genomes to identify promising DIP-STR candidates. After filtering for quality and variability, 23 new markers were validated through rigorous testing for sensitivity and specificity. These markers demonstrated their capability to detect minor DNA contributors in mixtures where the major-to-minor ratio exceeded 500:1, showcasing unparalleled precision. Additionally, Swiss population data was analyzed to confirm the markers' variability and applicability. The research proposes a panel of 30 DIP-STR markers for further development, with potential applications in both traditional forensic cases and non-invasive prenatal testing.
This study significantly enhances the forensic toolkit by providing a validated set of highly sensitive DIP-STR markers, capable of resolving complex DNA mixtures with unmatched accuracy. These findings not only improve current forensic methodologies but also lay the groundwork for advanced multiplex assays using both traditional and next-generation sequencing technologies. The development of these tools promises to refine forensic investigations, ensuring reliable DNA analysis in even the most challenging cases.
For more information, see the published paper.