Genomics evolution of an epidemic MRSA strain
Evolution of a hospital-acquired pathogen
Methicillin-resistant Staphylococcus aureus (MRSA) is a type of bacteria that is resistant to many commonly used antibiotics, and represents a major concern in healthcare settings. MRSA’s ability to adapt and resist treatment highlights the importance of strong infection control measures and ongoing research into bacterial evolution and antibiotic resistance.
Here we explored the genomic evolution of a specific MRSA strain known as ST228 SCCmec-I, that caused a major outbreak in a hospital setting over a decade ago. The research seeks to answer a critical question: what happens to such epidemic bacteria after an outbreak is controlled? By focusing on the genetic changes in the bacteria over time, this study provides essential insights into how pathogens adapt, evolve, and sometimes decline in their ability to spread. This understanding is vital for developing better infection control strategies, ensuring healthcare settings remain safe, and tackling the growing global issue of antibiotic resistance.

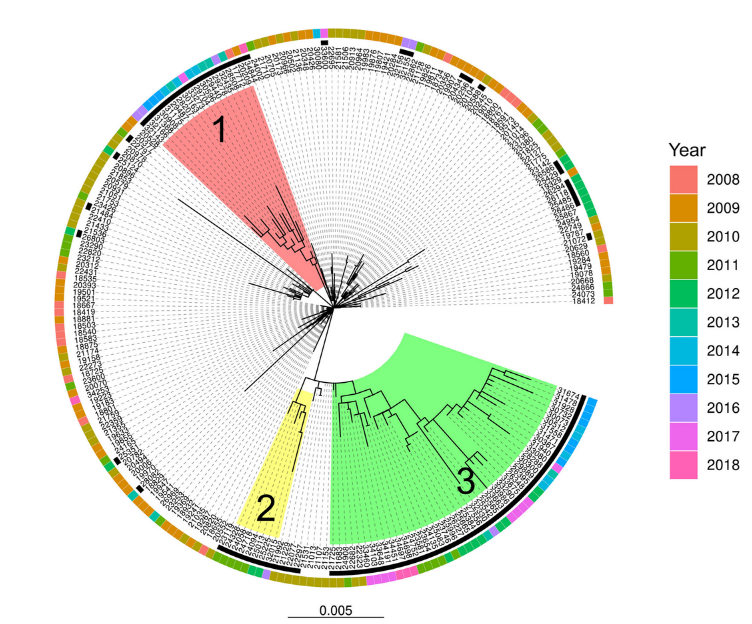
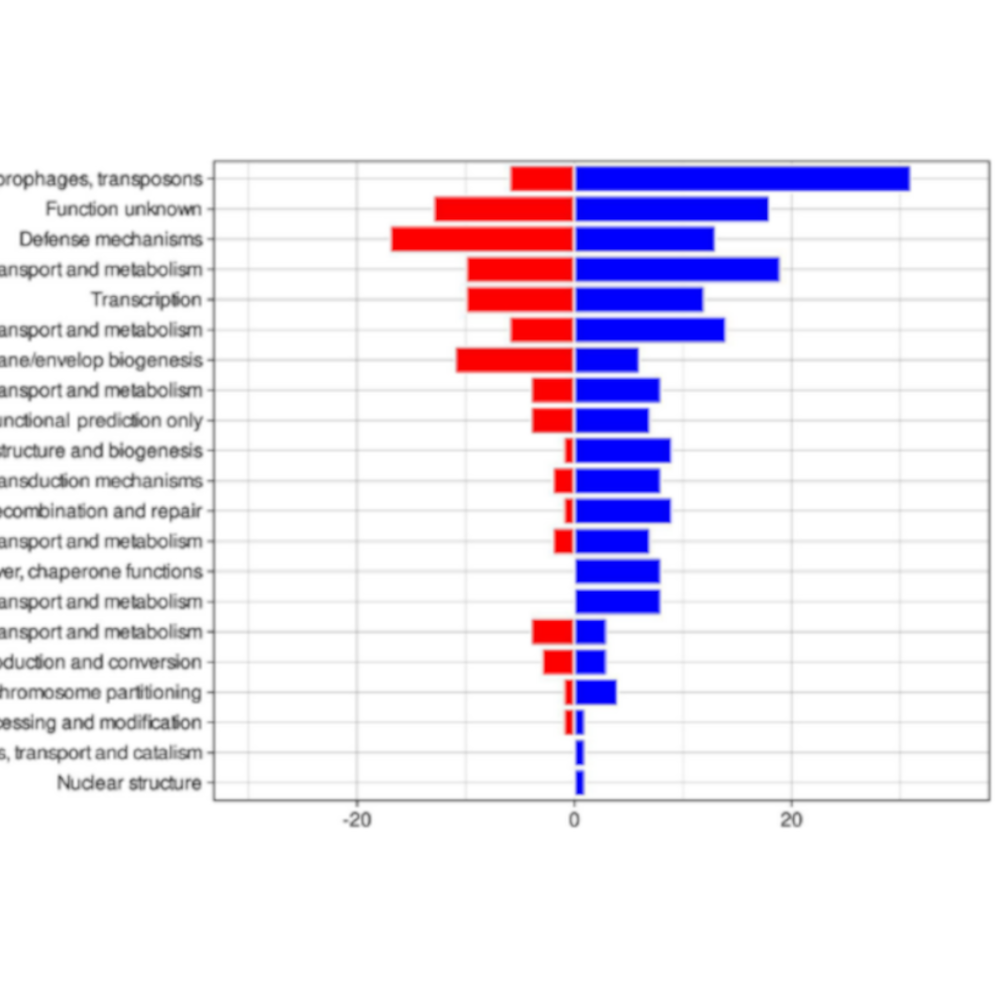
We collected and analyzed bacterial samples from patients over a 10-year period. Using whole-genome sequencing, we compared the DNA of MRSA isolates from the original outbreak to those collected in the years following it, allowing to track genetic changes in the strain. We found that the epidemic strain underwent significant genomic alterations, including the loss of genes associated with antibiotic resistance and virulence. At the same time, new genes were acquired, which appeared to favor long-term survival rather than rapid spread. GWAS and machine learning models identified genetic markers and features that were most associated with a decline in the strain’s ability to cause outbreaks. Interestingly, this decline in the bacteria’s epidemic potential coincided with the implementation of enhanced infection control measures, suggesting a combined effect of human intervention and bacterial evolution.
This underlined the importance of understanding how pathogens evolve over time, particularly in the context of antibiotic resistance and hospital outbreaks. The decline of the ST228 MRSA strain serves as a reminder that rigorous infection control measures can significantly reduce the spread of dangerous bacteria. However, the genetic changes observed in the strain also highlight the adaptability of pathogens to new environments, including those with heightened infection controls. By identifying specific genetic changes linked to the strain’s decreased epidemicity, the study provides a roadmap for future research and practical strategies to monitor, manage, and prevent bacterial outbreaks. These insights are especially relevant in the fight against antibiotic resistance, a pressing global health challenge.
For more information, see the published paper.